Published on 22.10.2025
Presentation
The Imagine Single-Cell platform will likely open during the second trimester of 2020 and will be located on the second floor of the Imagine institute.
We received funding from a sesame PIA, BPI region Ile de France call in 2019 to create a core facility specialized in gene expression and chromatin accessibility at the single-cell level. This platform will be open to both academics and industrials and will propose access to equipment, expertise, computational analyses as well as trainings in the single-cell field.
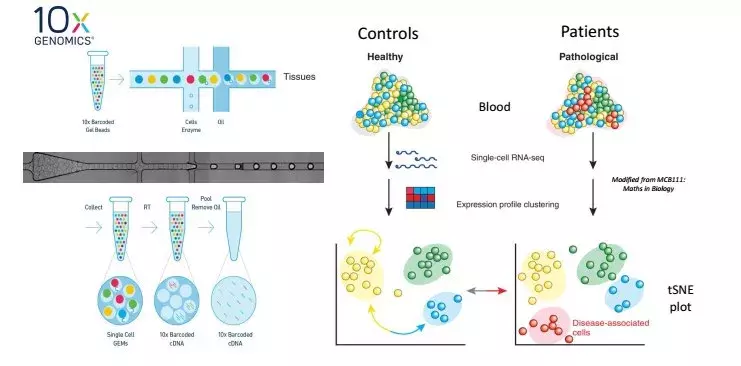
The RNA sequencing and chromatin accessibility, at the single cell level allow to analyze the difference in gene expression at a level never reached before. It helps to reveal rare populations of cells, extract molecular signatures, identify some unknowns process of regulation, follow trajectory of distinct cell lines under development or study the effect of local environment, stimulus or therapeutic molecules.
The Single Cell platform will be certified by 10X Genomics, one of the world leader company to deliver single-cell products and equipment.
Equipment
The single-cell platform will be equipped with :
- Chromium Controller in L1 facility , to generate single-cell encapsulation and libraries
- Chromium Controller in L3 facility , to generate single-cell encapsulation and libraries starting from cells infected by pathogens (HIV, Sars-Cov2 for example,…)
- Chromium Connect controller in L1 facility to perform single-cell transcriptomic libraries in an automated way
- PCR BioRad Thermocycler.
Services/applications proposed
Our services will go from consulting to set up an experiment, single-cell libraries generation, sequencing of libraries and computational analyses.
At the opening, the platform will propose all the techniques and protocols commercialized by 10X Genomics, to perform analyses of gene expression at the single-cell level
- 3’ Single Cell Transcriptomic
- 5’ Single Cell Transcriptomic + VDJ
- Feature Barcoding, CITE-seq protocol
- Single Cell ATAC-seq
- Spatial Transcriptomic
Single-Cell library generation and analyses
By using microfluidics, cells are individually isolated in water droplets. RNA of each cell is then extracted, reverse transcribed, barcoded into DNA, amplified, sequenced and analyzed. The results are shown as “t-SNE” where each cell is positioned in relation to the others according to its most expressed genes.
Team

Key numbers
1 cell per bead
10 000 Single-Cell librairies per sample
2000 most expressed genes analyzed per cell